Adaptation of SONAR-netCDF4 convention#
Echopype follows the ICES SONAR-netCDF4 convention ver.1 when possible. However, to fully leverage the power of label-aware manipulation provided by the xarray library and enhance coherence of data representation for scientific echosounders, the echopype developers have made decisions to deviate from the convention in key aspects.
Organization of multi-frequency data#
One important Echopype adaptation is the organization of multi-frequency data. Echopype implements a data structure that optimizes data access and filtering (“slicing”) efficiency and usability at the expense of potentially increased file storage.
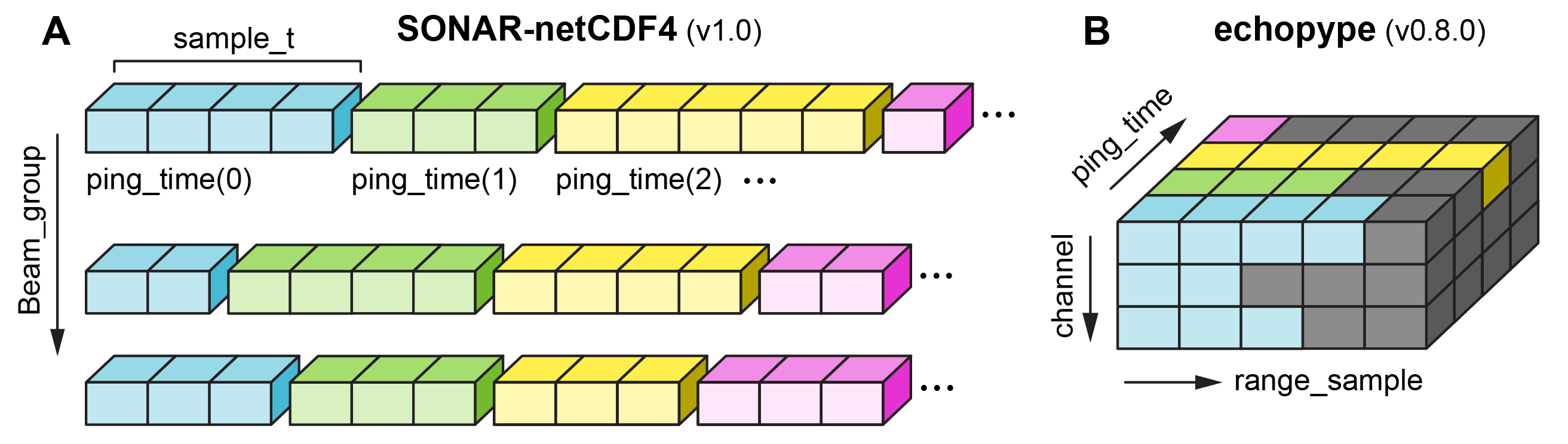
Specifically, the SONAR-netCDF4 convention defines that data variables, such as backscatter_r, from each sonar beam (i.e. frequency channel or transducers for typical scientific echosounder) are stored based on a one-dimensional ragged array structure that uses a custom variable-length vector data type (sample_t) and ping_time as its coordinate dimensions. In addition, each frequency channel is stored in a separate netCDF4 group (Sonar/Beam_group1, Sonar/Beam_group2, …).
Echopype restructures this multi-group ragged array representation into a single-group, 3-dimension ((channel, range_sample, ping_time)) or 4-dimensional ((channel, range_sample, ping_time, beam)) gridded representation across all channels. Here:
the
ping_timedimension follows the convention definitionthe
beamdimension, when exists, maps to the different sectors of split-beam transducersthe
channelandrange_sample(along-range sample number) dimensions are echopype-specific modifications
Data from each frequency channel are mapped along the channel dimension, and echo data from each ping are mapped along the range_sample dimension. These consolidated, uniform multi-channel (or multi-frequency) DataArrays are stored in Sonar/Beam_group1, Sonar/Beam_group2, and potentially other such groups (Sonar/Beam_group3, etc.) in the netCDF data model.
See Data from different echosounders for detail on core variables that store the echo data and the number of dimensions, which varies depending on the instrument setup.
NaN-padding#
Due to the flexibility in echosounder configuration, there can potentially be unequal number of samples along sonar range (i.e., length of the range_sample dimension) across different ping_time or channel. Echopype addresses this by padding NaN for pings or channels with fewer samples to maintain the uniform shape of a 3- or 4-dimensional gridded representation.
Below is a comparison of data representations defined in (A) the SONAR-netCDF4 convention and in (B) echopype, where the gray cells represent NaN-padded cells. This sketch illustrates the case of 3-dimensional gridded data such as backscatter_r from AZFP and EK60 data, or EK80 power/angle data.
Note
The NaN padding approach could consume large amount of memory in some specific cases due to the echosounder setup. This is an issue we are actively working on. See #1070 for detail.
Verifying compliance#
Ongoing echopype development creates a need to ensure that new modifications do not break the convention-based data structure unexpectedly, and that deliberate modifications are implemented consistently across instrument types. To assist with this need, we are developing a lightweight package that will verify the adherence of an EchoData object instance to the echopype adaptation of SONAR-netCDF4 version 1. The repository for this new, companion package, echopype-checker, currently contains a brief description of the package goals and operation as well as Jupyter notebooks that illustrate its use with specific raw data files.
