Development roadmap#
Scope and goals#
Echopype is a library aimed at enabling interoperability and scalability in processing ocean sonar data. The current focus and scope are on scientific echosounders widely used in fisheries and marine ecological surveys. We envision that Echopype will provide “building blocks” that can be strung together to construct data processing pipelines to bring raw data files collected by these instruments to “analysis-ready” data products that can be used by researchers in the ocean sciences community. We also plan for Echopype to be flexible in accommodating both local and cloud computing environments, such that data processing pipelines can be prototyped locally and scaled up for larger scale processing on the cloud.
To achieve these goals, we develop a workflow that focuses first on standardizing data to the widely supported netCDF data model, and based on the standardized data build computation and visualization routines by leveraging open-source libraries in the scientific Python software ecosystem, especially those in the Pandata stack.
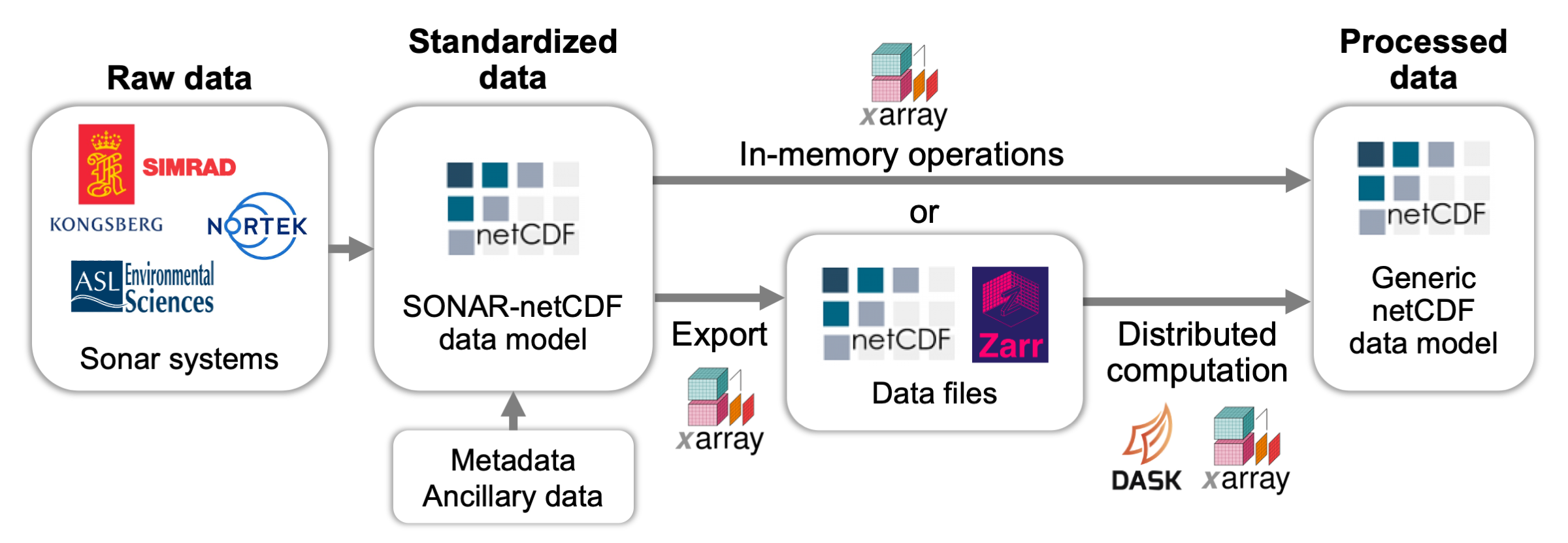
Data standardization#
Echopype contains a core data conversion and standardization subpackage that returns an EchoData object that allows easy and intuitive access and understanding of echosounder data in a netCDF data model. Currently Echopype converts raw instrument data files into netCDF4 or Zarr files following a modified version of the ICES SONAR-netCDF4 convention that we believe improves the data coherence and efficiency of data access (see details here). This conversion step ensures that downstream processing can be developed and executed in an instrument-agnostic manner, which is critical for tackling the tedious and labor-intensive data wrangling operations.
As the core data representation stabilizes across a few echosounder models, going forward we plan to:
Continue maintaining and updating the standardized
EchoDataobject in accordance with the evolution of community raw data conventionsEnhance adherence to community conventions of metadata and processed data, such as the ICES AcMeta, the new Gridded group introduced in SONAR-netCDF4 v2.0, and the Australia IMOS SOOP-BA conventions
Add support for data from other echosounder models, including historical data from Simrad EK/BI500 and the Biosonics DT4 files
Develop converter between data from major versions of Eechopype
Data processing levels and provenance#
In parallel with code development, the Echopype team is also working on defining “data processing levels” for echosounder data (see Echolevels). While no community agreement currently exists, these definitions are crucial for broader data usage, as evident in the widespread use of NASA satellite remote sensing data that are accompanied by a set of robust and well-articulated data processing level definitions.
In Echopype, currently many functions generate prototype data provenance and processing level information as data variables or attributes. Going forward we plan to:
Collaborate with the echosounder community to further refine the definitions for echosounder data processing levels
Improve data provenance preservation along the “chain” of data conversion and processing functions
Processing functions#
As the foundational data standardization components of Echopype mature, in the next stage of development we plan to redirect our attention to focus on expanding downstream data processing functionalities. The development will in principle follow the current subpackage grouping and prioritize the following items:
Commonly used, rule-based (i.e. non-ML) functions for noise removal, bottom detection, single target and swarm detection
Broadband echo processing, such as frequency-dependent Sv and TS computation
Calibration and utility functions, such as stadard target calibration procedure, updated estimates of sound speed and absorption coefficients, profile-based (rather than based on water-column averages) Sv and TS computation
Computational scalability#
Computational scalability is a core goal of Echopype development. We aim to provide scalable data processing capability for researchers both on their own personal computer and on computing clusters. The Echopype data conversion tools provide direct read/write interface with both local filesystems and cloud storage, and all downstream data processing functions also natively interface with both local and cloud resources through the combination of the Zarr, Xarray, Dask, and related libraries. However, we have found that the often irregular spacing and structure of echosounder data in time and space can impose substantial computational bottleneck and require custom optimization beyond stock Xarray functions to parallelize efficiently across computing agents. With a few important memory issues during data conversion resolved (see v0.8.0 release notes), going forward we plan to:
Benchmark data processing functions against diverse datasets of different volume (100s of GB to TB) and spatiotemporal features that can cause unintended memory expansion during computation
Leverage Dask delayed approaches and experiment with different Zarr chunking schemes to resolve computational bottlenecks
Companion developments#
Echopype focuses on data standardization, aggregations, and processing for building efficient and scalable data workflow. To address other needs in integrative analysis of echosounder data, the Echopype team is taking a modularized approach to develop the following companion libraries:
Echoflow: Orchestrate workflow on the cloud or local platforms
Echoshader: Interactive visualization widgets leveraging the HoloViz suite of tools
Echolevels: Proposed specifications of echosounder data processing levels
Echoregions: Interface with echogram interpretation masks from physics-based or data-driven methods
EchoPro: Incorporate net-based data and scattering models for biomass estimation, currently focused on Pacific hake
